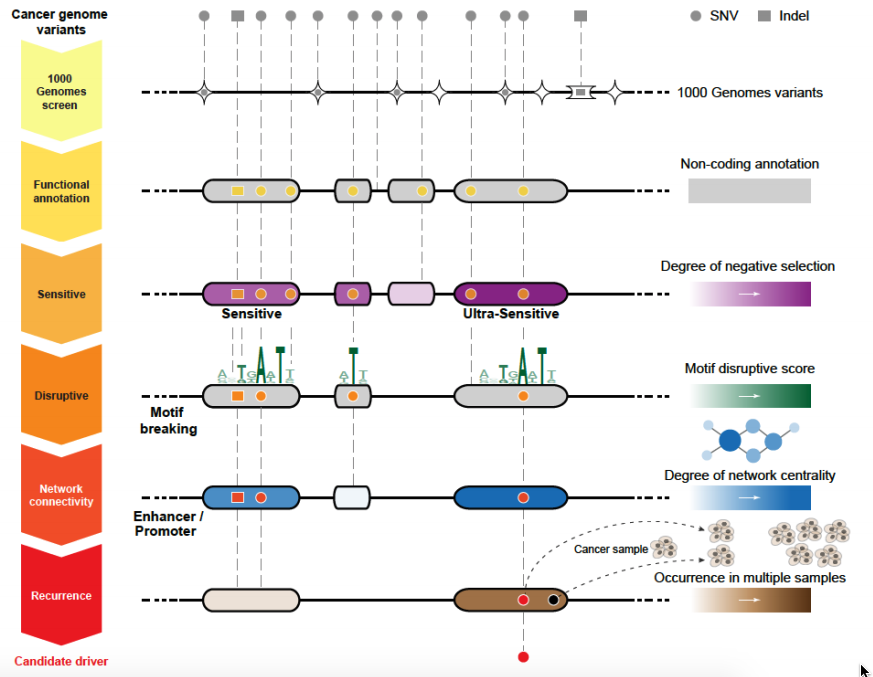
Funseq
Funseq/Funseq2 is specialized to prioritize somatic variants from cancer whole genome sequencing.
Keywordssomatic variants weighted scoring systemcoding non-coding
Availabilitygithub

Funseq
Funseq/Funseq2 is specialized to prioritize somatic variants from cancer whole genome sequencing.
Keywordssomatic variants weighted scoring systemcoding non-coding
Availabilitygithub

LARVA
LARVA is a computational framework designed to addresses issues that have made it difficult to derive an accurate model of the background mutation rates of noncoding elements in cancer genomes: limited noncoding functional annotation, great mutation heterogeneity, and potential mutation correlations between neighboring sites.
Keywordsmutation burden beta-binomial distributionreplication timing non-coding
Availabilitygithub

MOAT
MOAT (Mutations Overburdening Annotations Tool) is a computational system for identifying significant mutation burdens in genomic elements with an empirical, nonparametric method. MOAT offers two types of permutation algorithm: one that permutes the locations of annotations (MOAT-a), and one that permutes the locations of variants (MOAT-v).
KeywordsMutation burden permutationempirical nonparametric
AvailabilityWebserver

GRAM
GRAM is a generalized model built to incorporate multiple genomic predictors in order to predict a well-defined experimental target: the expression-modulating effect of a non-coding variant in a cell-specific manner.
Keywords cell specificfine-mapping non-coding
Availabilitygithub

GenoDock
GenoDock employs a supervised learning model using the pseudo gold-standard set as a target feature. Its goal is to predict the sensitivity of drug binding to personal SVs.
KeywordsSupervised learning weighted scoring systemcoding Drug binding
Availabilitywebserver

ALoFT
ALoFT allows users to annotate and predict the disease-causing potential of loss of function (LoF) variants.
Keywordsloss of function domain annotationevolutionary conservation biological networks
Availabilitywebserver

VAT
VAT (Variant Annotation Tool) was developed to functionally annotate variants from multiple personal genomes at the transcript levels as well as to obtain summary statistics across genes and individuals.
Keywordsvariant annotation allele frequencycoding genotype data
Availabilitywebserver

RADAR
RADAR is a variant-socring framework based on the integration of the ENCODE-RBP experiment portfolio.
KeywordsENCODE-RBP weighted scoring systemTissue-specific Somatic and germline
Availabilitywebserver